Home | Introduction | Oxygen Binding | F Helix Transition | Quarternary Shift | Synopsis | Bibliography
The hemoglobin molecule resembles a finely tooled mechanism that has very little slop. The binding of O2 requires a series of tightly coordinated movements:

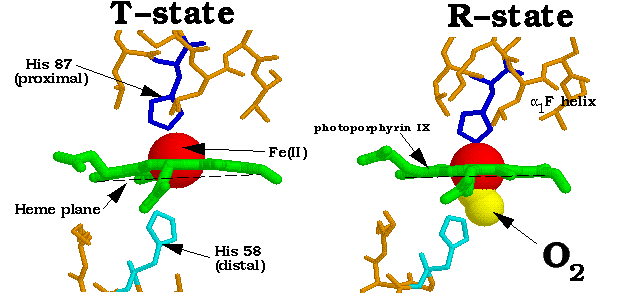
1. The Fe(II) of any subunit cannot move into its heme plane without the reorientation of its proximal His so as to prevent this residue from bumping into the porphyrin ring.
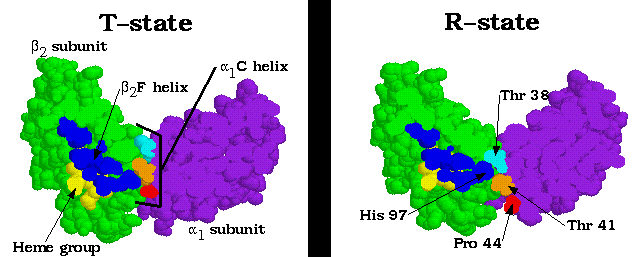 2. The proximal His is so tightly packed by its
surrounding groups that it cannot reorient unless this movement is accompanied
by the previously described translation of the F helix across the heme plane.
2. The proximal His is so tightly packed by its
surrounding groups that it cannot reorient unless this movement is accompanied
by the previously described translation of the F helix across the heme plane.
3. The F helix translation is only possible in concert with the quarternary shift that steps the alpha1C-beta2FG contact one turn along the alpha1 C helix.
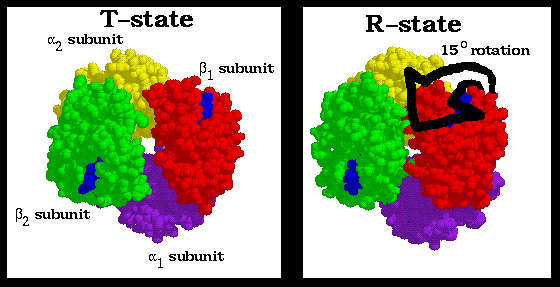
4. The inflexibility of the alpha1-beta1 and alpha2-beta2 interfaces requires that this shift simultaneously occur at both the alpha1-beta2 and alpha2-beta1 interfaces.
Consequently, no one subunit or dimer can greatly change its conformation independently of the others. Indeed, the two stable positions of the alpha1C-beta2FG contact limit the Hb molecule to only two quarternary forms, R and T.
The t state has reduced oxygen affinity, most probably because its Fe-O2 bond is stretched beyond its normal length by the steric repulsions between the heme and the oxygen . . . Unliganded subunits in the r state have an increased oxygen affinity because they are already in the O2-binding conformation.
A final note: because this mechanism (known as the Perutz mechanism) is largely based on the static, crystal structures of the T and R end states, it should be considered at least partly conjectural. However, it is the most widely accepted hypothesis.
All the material in this tutorial was taken from Voet, Judith G. and Voet, Donald, Biochemistry. John Wiley and Sons, Inc. New York, NY. 1995. Chapter 9: Hemoglobin: Protein Function in Microcosm
All protein crystal data was taken from the BrookHaven Protein Database and graphically depicted using Rasmol, a shareware program by Roger Sayle. The images in this tutorial were edited using Showcase on a Silicon Graphics machine, as well as Photoshop and Graphics Converter on Macintosh. The web site was created using Adobe's PageMill.